Research Areas
In recent years we have worked on projects in the following areas:
- Microbial Mats in Yellowstone Hotsprings
- Drivers of Genetic Diversity in the Marine Microbiome
- Eco-Evolutionary Dynamics of Diverse Communities
- Cancer Biology
- Population Genetics and Evolutionary Dynamics
- Experimental Evolution of Microbes
- Theoretical Neuroscience
If you're interested in learning more, contact the appropriate researcher.
Microbial Mats in Yellowstone Hotsprings

With collaborator Devaki Bhaya at the Carnegie Department of Plant Biology we are interested in understanding the patterns of genetic diversity found in naturally occurring communities of the thermophilic cyanobacteria Synechococcus that live in the Yellowstone caldera. How important are recombination and selection in these natural microbial populations, and can we observe their signatures in data? Since we are particularly interested in fine-scale diversity, we have been working with a collection of single cell sequences, which give us high resolution and the power to assess recombination between individual cells.
Drivers of Genetic Diversity in the Marine Microbiome
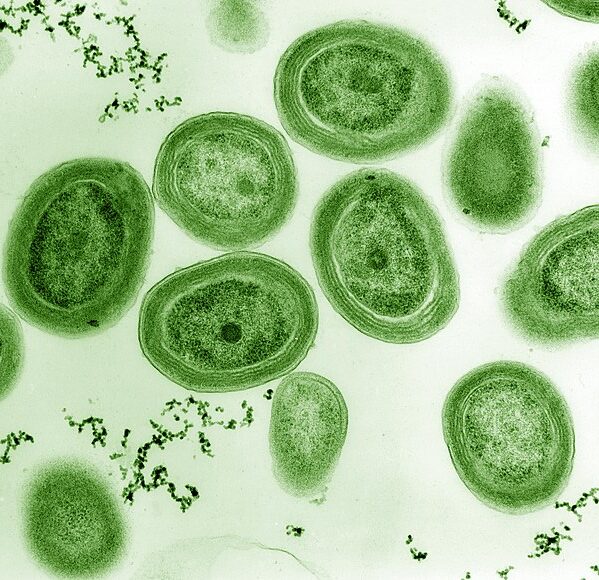
Prochlorococcus is an ocean-dwelling bacterium that is responsible for 10% of global carbon fixation. It is the most abundant photosynthetic species on Earth and is a highly diverse. Although Prochlorococcus is critical for supporting life as we know it, little is known about how this species and its various subclades evolve and interact, particularly in terms of recombination — a process in which bacteria exchange genes with their contemporaries, making it such that each gene in a cell can have a different “parent.” Recombination is typically thought to occur between closely-related cells, but by studying the genomes of hundreds of individual Prochlorococcus cells we hope to obtain a deeper understanding of how recombination structures genetic diversity, from the most closely-related cells found to the most distantly-related.
More generally we are interested in looking for signatures of genetic exchange between microbes in nature, to see whether these can explain the bewildering diversity that seems to be present across many orders of magnitude in genetic divergence.
Eco-Evolutionary Dynamics of Diverse Communities
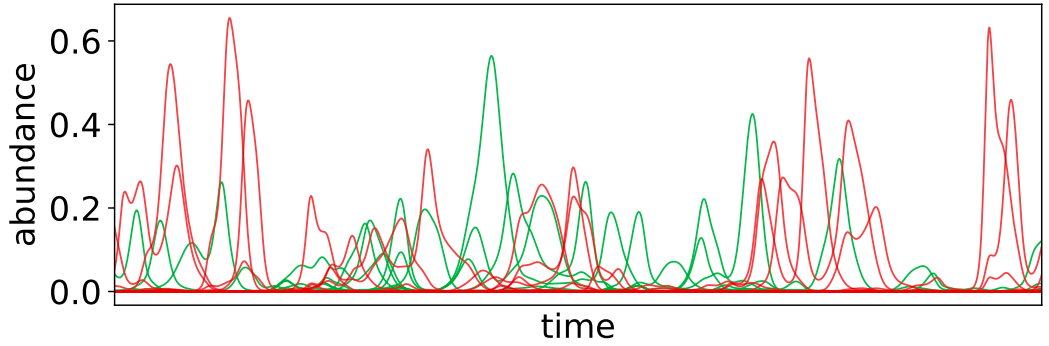
How do ecosystems maintain diversity? What does evolution look like when inter-species interactions are important? What is the co-evolutionary dynamics of hosts and pathogens? How do underlying ecological dynamics impact the evolutionary trajectories of communities? Even basic questions about evolutionary processes in complex ecosystems are hard to answer. Yet, systems with multiple interacting organisms are ubiquitous in nature; for example, most microbial species cannot grow in isolation in laboratory conditions. We are working to lay the foundations for eco-evolutionary theory using a combination of mathematical modeling, simulation, and data analysis. We are particularly interested in understanding the interplay between the various dynamical processes that are important in ecological systems, and exploring whether or not they can give rise to the phenomena we observe in nature. It is an especially important time to work on these theoretical problems, in order to make sense of the vast amounts of data currently being collected in microbial ecological systems from ocean-dwelling cyanobacteria to the gut microbiome. Current research directions include:
- Bacteria-phage coevolution
- Resource-mediated interactions
- Role of spatial structure in stabilizing diversity
Cancer Biology
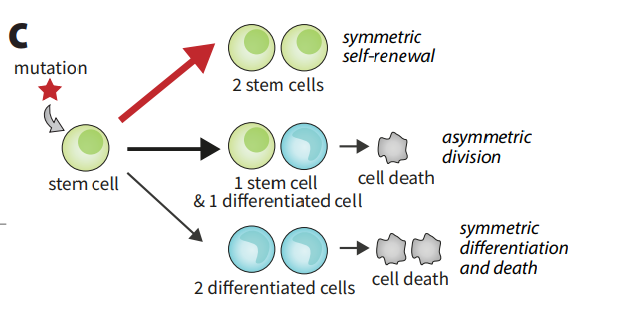
Cancer is fundamentally an evolutionary disease, and advancements in genomics have made it possible to study it as such. There are many unanswered dynamical questions. How does cancer arise and progress? What are the best methods of early detection? Is it possible to understand, model, and predict the behaviors of cell lineages?
Blood cancers may be particularly amenable to theory-inspired approaches, in part due to their (relatively) simple spatial structure. There are many mutations that are commonly detected in the blood cells of leukemia patients; these ‘driver mutations’ appear associated with blood cancer development. Many of these mutations are present at substantial frequencies in the blood of healthy individuals and in fact may be nearly ubiquitous in healthy adults. Our modeling work attempts to understand the importance of the distribution of these driver mutations by constructing simple evolutionary models and using them to ask quantitative questions about data. Our goal is to use our insights to understand the progression of the disease, offer pragmatic advice about sequencing-based detection, and develop frameworks that can be used to address specific clinical and biological questions.
Population Genetics and Evolutionary Dynamics
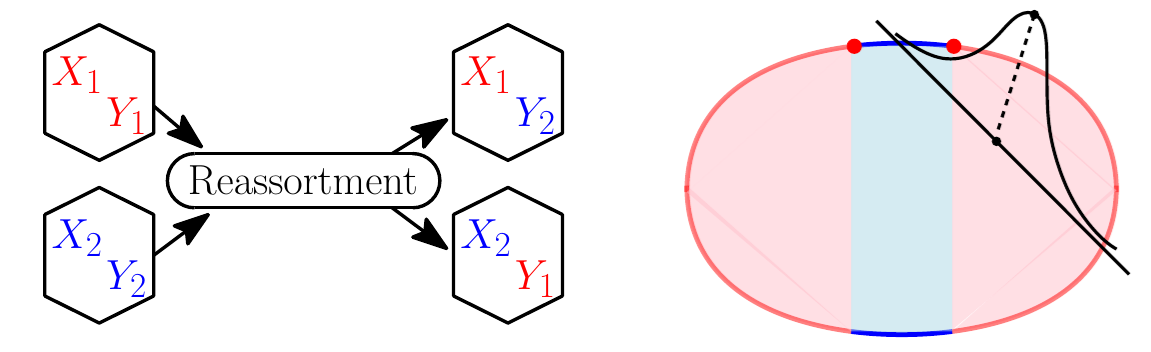
What are the relevant processes in the dynamics of evolution? What are their timescales, and how do they relate to the microscopic parameters in simple models? We study a variety of basic theoretical models of evolution, using analytical arguments augmented with computational exploration to develop intuition for the statistical dynamics of evolution.
Emphasis is placed on gaining an understanding which transcends the specifics of a model, and ideally helps guide the development of which observables are important for understanding real populations as well. Our use of asymptotics takes advantage of the area where humans still have an advantage over computer models and simulation: extrapolation over a wide range of parameter space. Recent projects include:
- Two chromosome model, intermediate recombination
- Epistasis on high dimensional random landscapes
Experimental Evolution of Microbes
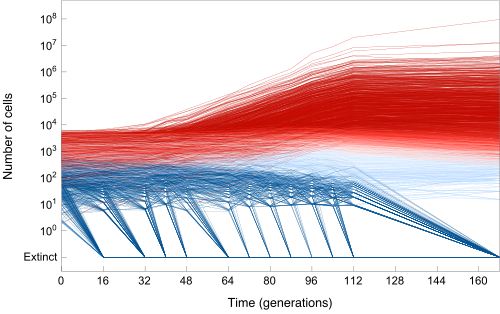
The evolution of large asexual cell populations underlies 30% of deaths worldwide, including those caused by bacteria, fungi, parasites, and cancer. However, the dynamics underlying these evolutionary processes are poorly understood because they typically involve many competing beneficial lineages. We helped to develop a sequencing-based ultra high-resolution lineage tracking system and associated quantitative models and analysis tools to track >500,000 cell lineages simultaneously. The central goal of this research drive is to develop a predictive understanding of how rapidly cell populations adapt, and specifically what factors matter most (e.g. population size, mutation rate, growth rate, drug concentration, recombination rate etc). Understanding how these parameters affect the speed of evolution could lead to improved treatments for evolutionary diseases such as bacterial infections, cancer and HIV.
Theoretical Neuroscience
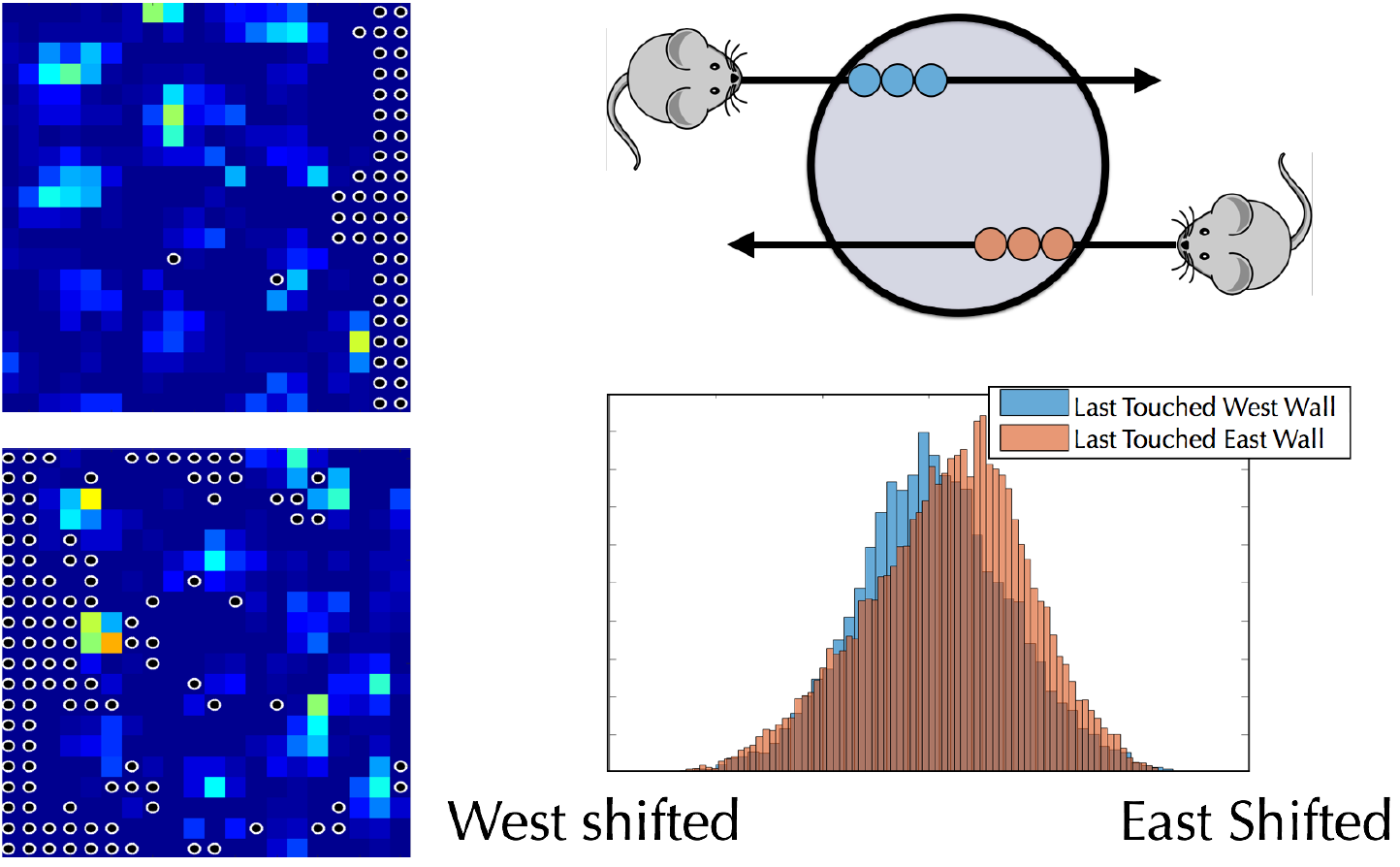
In order to navigate, animals need to maintain a representation of their environment as well as their position within it. How are these representations learned and encoded, and how do they interact with each other? Relatively recently, “grid neurons”, with a characteristic hexagonal firing pattern, have been discovered in mammals. In collaboration with the Ganguli and Giocomo labs, we work on a combination of theory and data analysis to understand how an animal navigates and learns about its environment from the combination of step-counting and landmarks.