- HOME
- RESEARCH
- PUBLICATIONS
- PATENTS
- CONTACT
Bioseparations
With the development of “next-gen” sequencing technologies including pyrosequencing, sequencing-by-synthesis, and single-molecule sequencing, the acquisition of large amounts of sequence data in a massively parallel fashion is becoming reasonable. However, next-generation technologies cannot yet routinely provide 600+ bases of contiguous high-accuracy DNA sequence, as needed to sequence an entire human exon accurately in a single pass.
Also, these technologies are not capable of providing the long, accurate reads needed to reassemble repeat-rich loci that occur very frequently in human genomes. Therefore, DNA electrophoresis will continue to be a uniquely useful technology for certain applications, such as the de novo sequencing and assembly of novel, large, complex genomes, and the medical sequencing of a key gene regions as will needed in future clinical practice. Medical and diagnostic applications of the future could benefit from the availability of inexpensive, disposable, all-in-one devices for DNA extraction, amplification and separation/analysis on a microchip electrophoresis format. Microfluidic chip-based DNA separations can be at least 10 times faster, and also should be less expensive than capillary array electrophoresis.
The bioseparations group in the Barron laboratory specializes in developing novel polymeric materials and comprehensive strategies and procedures to enable efficient electrophoretic separations of DNA for four-color sequencing and genetic mutation detection applications. These projects involve the synthesis, design, purification, and physical characterization of novel water-soluble polymers and nanogels, and their testing for DNA separations on microfluidic devices. We have projects that focus on the development of optimal entangled polymer networks for ultra-fast DNA sequencing and genotyping on chips. We are also developing a free-solution method of electrophoretic DNA sequencing, utilizing de novo-designed protein and poly-N-methoxyglycine “drag-tags” that introduce a strong size-dependence to the electrophoretic mobilities of biomolecules in free solution, high-field electrophoresis on microfluidic devices. Our integrated microdevices team is working to design, fabricate, and evaluate a variety of novel microfluidic devices that, when used with our novel polymer wall coatings, separation networks, and drag-tags, can give extremely fast (< 7-minute), high-accuracy sequencing of long regions of contiguous DNA sequence.
Ultrafast DNA Separations by High-Field Microchannel Electrophoresis
DNA Sequencing
DNA Sequencing
The transition from traditional slab gel electrophoresis to microchannel electrophoresis (i.e. capillary or microchips) allows for a fast, cost-effective genetic analysis. Traditional slab gel electrophoresis, as still heavily used by molecular and cellular biologists, employs highly crosslinked polymer gels to separate DNA based on size or conformation. However, these same crosslinked polymers are highly viscous and ultimately pose problems for the rapid microchannel loading desired for high-throughput DNA analysis. An attractive alternative to crosslinked polymer gels is uncrosslinked polymer solutions, which allow facile loading and unloading due to their shear thinning rheology. Above appropriate concentrations and in the absence of shear stress, uncrosslinked polymers form physically entangled networks that provide the “obstacle course” that is necessary to accomplish the size-based separation of DNA molecules by electrophoresis. Our lab focuses on creating optimal sieving matrices for DNA samples of different types, by developing polymers and nanoscale hydrogel beads with designed, controlled chemical compositions and physical properties. (see Fredlake CP, Hert DG, Kan CW, Chiesl TN, Root BE, Forster RE, Barron AE, "Ultra-fast DNA sequencing on a microchip by a hybrid separation mechanism that gives 600 bases in 6.5 minutes," PNAS 2008, 105: 476-481.)
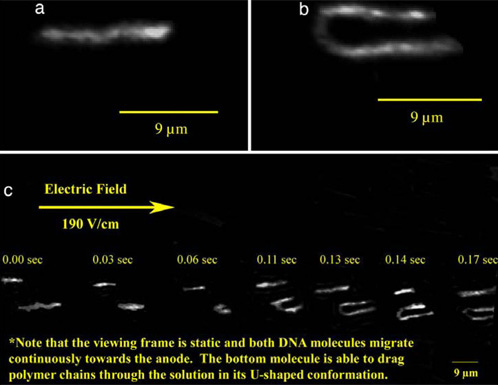
Figure: Images captured from DNA imaging videos. (a) λ-DNA is reptating through the polymer network. (b) This is an image of a λ-DNA molecule that has hooked around the polymer matrix in a U-shaped conformation and is dragging the disentangled matrix polymers. (c) A series of frames at the shown time intervals show two DNA molecules moving through the network (same molecules in a and b). The top molecule reptates through the entire viewing frame in the given time. The lower molecule is reptating at first and then hooks and drags the polymer network through the viewing frame.
Genetic Screening
Traditionally, single-stranded conformation polymorphism (SSCP) and heteroduplex analysis (HA) genetic mutation screening have involved the heat-denaturation of a sample of pure, diluted, double-stranded PCR products and rapid cooling, followed by slab gel or capillary electrophoresis under non-denaturing conditions in a highly resolving separation matrix. During cooling in dilute conditions, some single-stranded DNA fragments do not find their complement, but instead collapse and fold into complex three-dimensional shapes dictated by their primary sequence. Other single-stranded DNA fragments form duplexes consisting of perfectly complementary strands (homoduplexes) or duplexes containing base mismatches (heteroduplexes). These differently folded or hybridized forms of DNA often can be distinguished by electrophoresis as separated zones or peaks, in order to detect differences in DNA sequence between a patient sample and "wild-type" sample. This can be used for a rapid screen, since for many genes, sequence alterations are relatively rare, and one might not wish to fully sequence each gene region (for example, for the p53 gene, which is more than 20,000 DNA bases in length, split over 11 exons with intervening introns). Our group is working to develop advanced polymer materials and comprehensive protocols and devices needed for clinical implementation of our novel tandem SSCP/HA assay on microchips, to rapidly screen certain gene regions for sequence differences from the wild-type gene (see Christa N. Hestekin, John P. Jakupciak, Thomas N. Chiesl, Cheuk Wai Kan, Catherine D. O'Connell, Annelise E. Barron, "An optimized microchip electrophoresis system for mutation detection by tandem SSCP and heteroduplex analysis for p53 gene exons 5-9", Electrophoresis, 2006, 27, 3823-3835).
Single-strand conformation polymorphism (SSCP)
Single-strand DNA fragments adopt folded conformations that maximize the number of intra-strand base pairs formed, and have irregular three-dimensional shapes. Ideally, differences in three-dimensional shapes of mutant and wild-type single-stranded DNA conformers will result in different electrophoretic migration patterns, which may be detected and utilized for identifying DNA specimens containing mutations.
Heteroduplex analysis (HA)
Double-strand DNA mismatches (i.e., the formation of a heteroduplex, when mutant and wild-type DNA molecules are mixed, denatured, and then allowed to renature) can cause the formation of unusual secondary structures including "bubbled" regions that break the usual double helix form of DNA. Similar to SSCP analysis, the presence of mismatches may be detected during electrophoresis. Both SSCP and HA require the use of a highly entangled, molecularly confining DNA separation matrix that can reproducibly separate single-stranded DNA conformers that are formed during renaturation.
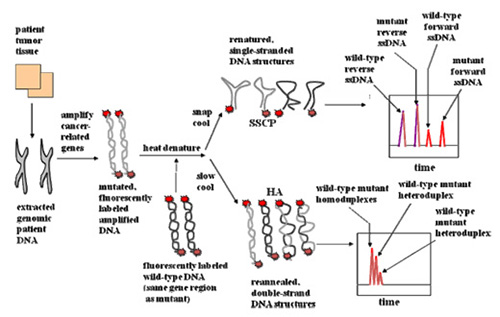
Figure: Schematic of SSCP and HA used for rapid screening for sequence differences between wild-type and patient genes.
- Start with source of DNA such as a patient tumor.
- Since mobility shift assay, must combine with wild-type or normal DNA.
- For SSCP, rapidly cool samples so that the DNA fold on themselves in unique conformations depending on the sequence. These conformational differences allow mutation detection as shown in idealized manner.
- For HA, cool more slowly to allow DNA to reanneal; this allows reformation of some of the original wild-type and mutant homoduplexes, but also allows formation of heteroduplexes, which occur when a wild-type and mutant anneal to each other. Due to the differences in sequence, the heteroduplexes will contain conformational differences that allow them to be seperated from the homoduplexes as shown in idealized manner.
Free-Solution Conjugate Electrophoresis (FSCE)
Traditional electrophoresis in polymeric sieving matrices has several drawbacks, however, including a limited ability to separate large DNA molecules, poor performance under high electric fields, and practical difficulties with engineering microdevices for loading viscous polymer into microchannels. The Barron group has developed a new method, Free-Solution Conjugate Electrophoresis (FSCE), which allows size-separation of DNA in the absence of a sieving matrix. In FSCE, each DNA molecule in a sample is covalently attached to a unique frictional modifier, or “drag-tag,” that serves to modify the free-solution electrophoretic mobilities of the DNA in a size-dependent manner by breaking the linear scaling of the charge-to-friction ratio. We have recently demonstrated the use of a first-generation, genetically engineered protein drag-tag for four-color sequencing of ~180 bases of DNA by capillary electrophoresis in less than 15 minutes, in the absence of a sieving matrix. Chip-based separations of these DNA sequencing fragments conjugated to the protein drag-tags have been performed and produce short reads, presently limited by drag-tag size and the sample injection method. We have also shown the capability of FSCE in genotyping assays such as single-base extension for multiplex detection of up to 16 p53 mutations. We believe that FSCE may be the key technological breakthrough that enables rapid, high-throughput sequencing and genotyping on automated, integrated microfluidic devices.
DNA Sequencing by Free Solution Conjugate Electrophoresis
One focus of our research has been the use FSCE for DNA sequencing in free solution. The basic approach of DNA sequencing by FSCE is to attach an uncharged, monodisperse perturbing entity (a "drag-tag") to each molecule of DNA, creating hydrodynamic drag which is not related to DNA size and hence allows the separation of differently sized DNA molecules. If each DNA molecule in a mixture is conjugated to the same monodisperse drag-tag, the mobility of small DNA molecules is reduced more than the mobility of large DNA molecules, and separation based on size is achieved.
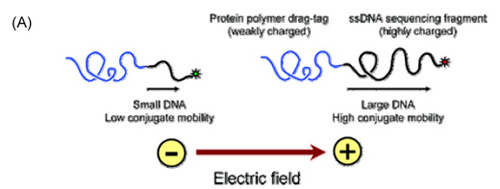
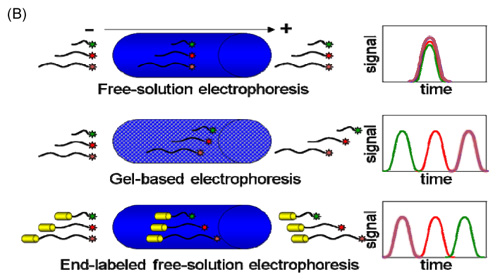
Figure: (A) Schematic illustration of end-labeled free-solution electrophoresis of DNA sequencing fragments. Each ssDNA fragment is conjugated to an identical, neutral (or nearly neutral) protein polymer drag-tag, which is present during the Sanger cycle sequencing reaction and then adds additional friction during free-solution microchannel electrophoresis. The additional drag impacts the mobility of DNA fragments inversely according to size, such that conjugates comprising large DNA fragments migrate close to the free-solution mobility of DNA, while conjugates with smaller DNA are slowed down substantially, according to their size. (B) Comparison of free-solution electrophoresis with no DNA separation to size-based separation using either gel-based electrophoresis or ELFSE. Note that the larger fragments elute first in ELFSE, the reverse of the gel-based separation.
A primary research effort has been the design of a suitable drag-tag for DNA sequencing. To achieve adequate analyte resolution and unambiguous sequencing results, an ideal drag-tag should have the following characteristics:
- Should be large (>250mer), adding a significant amount of hydrodynamic drag to DNA
- Must be absolutely monodisperse
- Must be suitable for attachment to a DNA molecule at a unique point, typically the terminus
- Should have minimal non-specific interactions with DNA or with the walls of the capillary or microchannel separation device
To be practical, the tag should also be stable, easy to synthesize, and easily conjugated to DNA. Polypeptoids and peptide/peptoid chimerae appear to be promising candidates for drag-tags, as they meet all of these characteristics. Synthetic polypeptoids, naturally occurring polypeptides, and engineered polypeptides have all been studied by our group. (see Meagher RJ, Won JI, Coyne JA, Lin J, Barron AE, "Sequencing of DNA by free-solution capillary electrophoresis using a genetically engineered protein polymer drag-tag", Anal. Chem. 2008, 80, 2842-2848.)
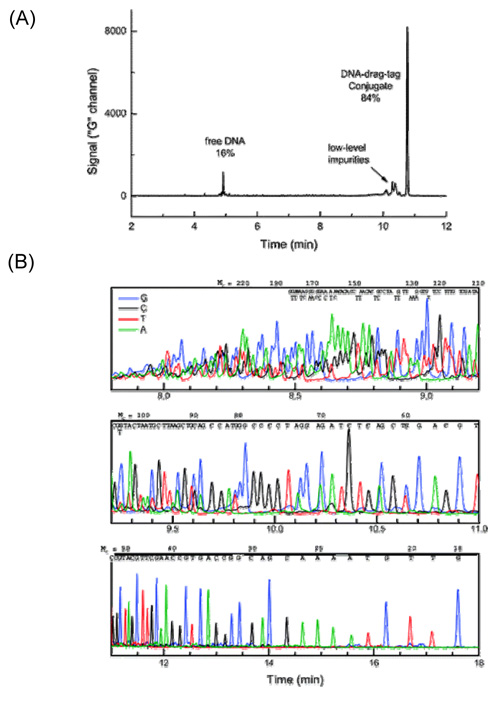
Figure: (A) Electrophoretic analysis of single-base extension products from the protein polymer drag-tag conjugated to a 5‘-thiolated 17-base M13mp18 sequencing primer. The analysis shows the monodispersity of the drag-tag and the efficiency of conjugation of the thiolated primer. (B) Four-color electropherograms for the products of a sequencing reaction carried out using the 127mer protein polymer drag-tag-labeled M13 primer and the SNaPshot SBE kit with dNTPs at a total concentration of 800 μM in the sequencing reaction.