
HUMAN FOAMY VIRUS (HFV)
 |
HUMAN FOAMY VIRUS (HFV)
|
HFV belongs to the genus of spumaviruses. Although it has been isolated
from patients with various neoplastic and degenerative diseases such
as myasthenia
gravis, multiple sclerosis, thyroditis de Quervain and Graves’ disease,
an etiological role for the virus has yet to be identified and little is known
about its prevalence in human populations. There is also a lot of controversy
about the criteria used to identify the isolate as HFV in most of these early
studies, and so it may be that HFV is really an orphan virus and is just coincidentally
associated with these disorders.
Like other Foamy Viruses, HFV has many characteristics that set it apart
from the other well-known retroviruses, including a distinct morphology,
budding
from the endoplasmic reticulum rather than the plasma membrane and a unique
replication strategy.
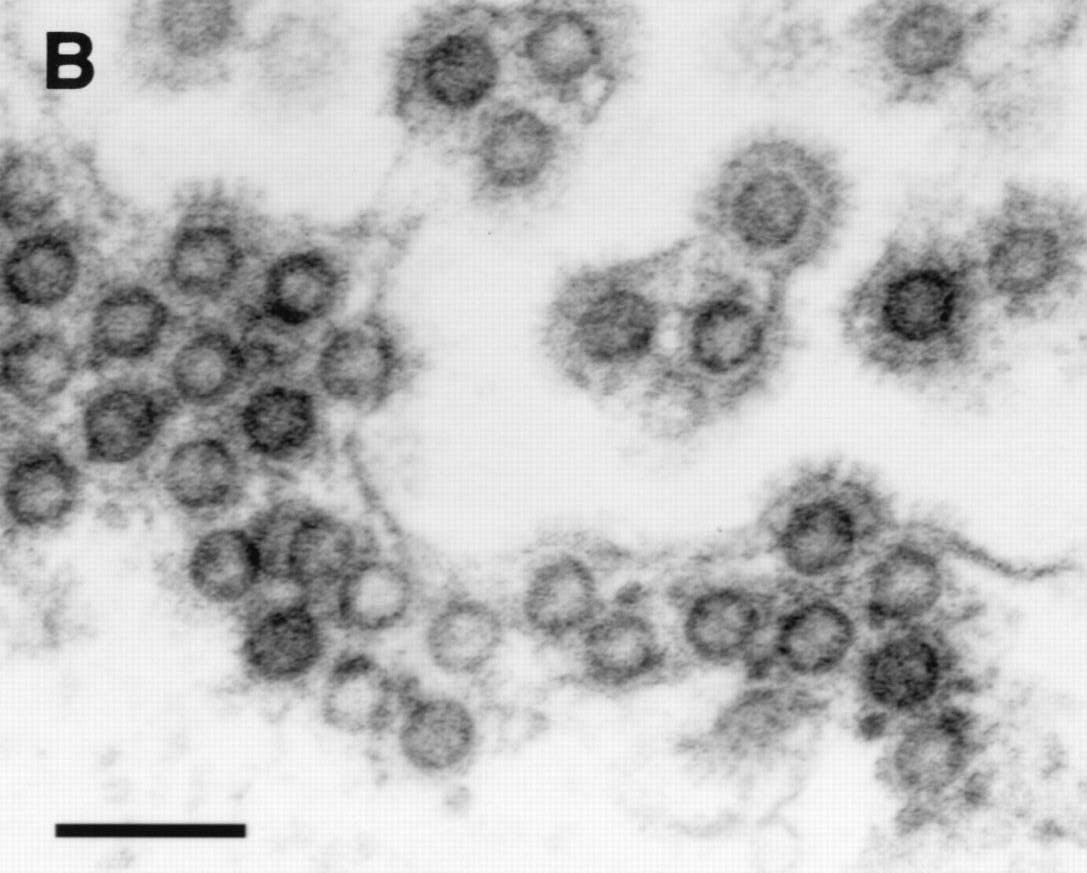
HFV particles shown budding from the ER (note immature particle appearance and characteristic envelope spikes)
Foamy Viruses
are considered complex retroviruses because they encode viral proteins that
are not incorporated into viral particles. The HFV genome encodes the canonical
retroviral gag, pol, and env genes, as well as at least two additional genes
termed tas (bel-1) and bet. Although a number of putative functions have
been proposed for the bet gene, its role in vivo is still unknown. The tas
gene (transactivator of spumavirus) is however, known to be required for
viral replication. It encodes a protein that transactivates a long terminal
repeat (LTR) promoter, which remains transcriptionally silent in the absence
of Tas. HFV is also known to have a second promoter, termed the internal
promoter (IP) located within the env gene, which drives tas and bet gene
expression and is also transactivated by Tas protein. During the early stages
of infection, the IP directs the synthesis of tas and bet mRNAs. Once the
Tas protein is synthesized, transcription begins from the LTR, leading to
the accumulation of gag, pol, and env mRNAs and ultimately to new viral particles.
The retroviral structural genes also behave in a distinctive way in HFV.
The virus particle has an immature appearance because the Gag protein
is not efficiently
cleaved into the mature viral proteins seen in most other retroviruses.
The Pol precursor protein is only partially cleaved, and after protease
cleaves
the integrase domain, it leaves the protease and reverse transcriptase
domains in the same molecule. The Env protein is cleaved into surface
and transmembrane domains, as in other retroviruses, but it contains
an endoplasmic reticulum retention signal as a result of which HFV has
the distinction of budding from the ER.
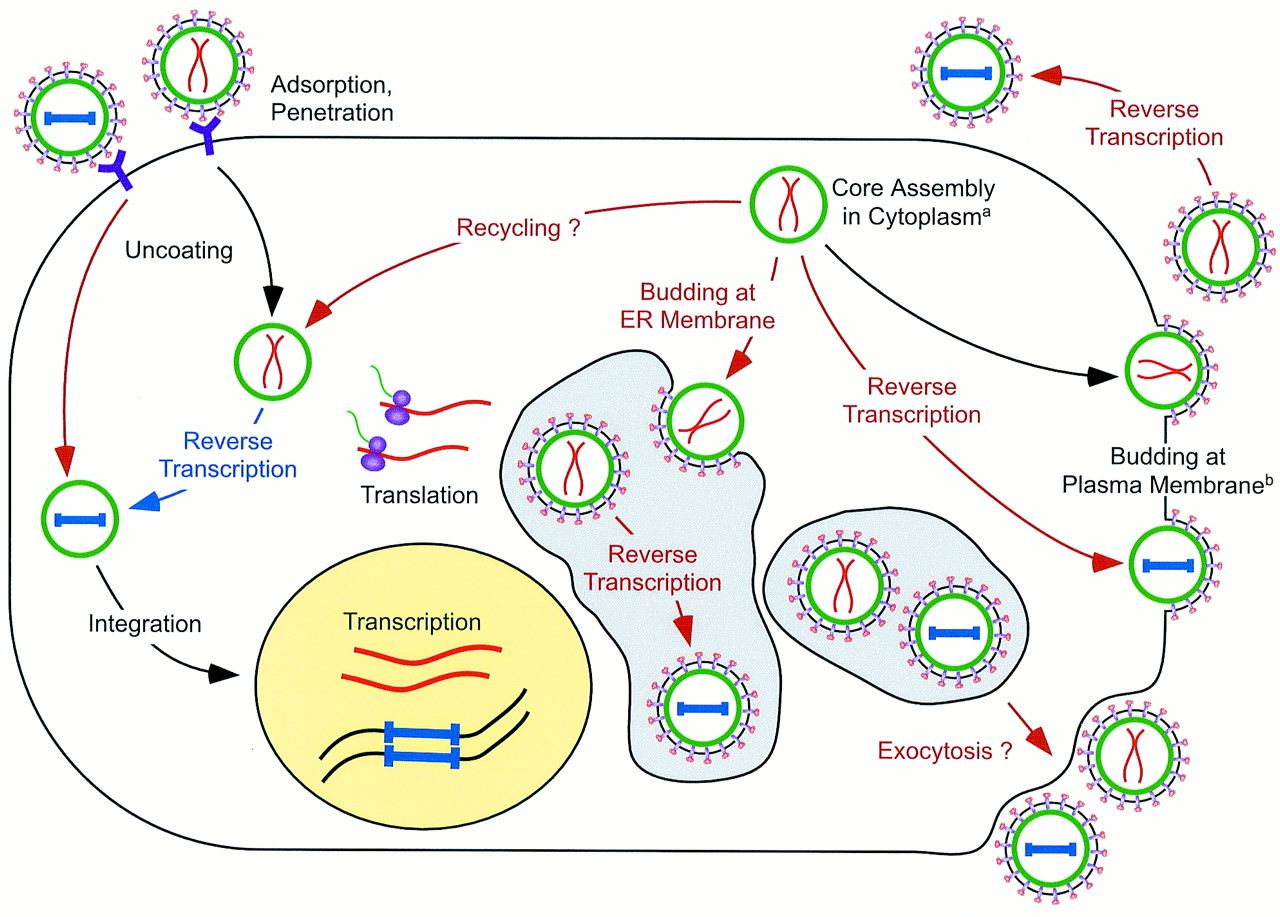
HFV Life Cycle
HFV replication also differs from conventional retroviruses, and in many ways resembles that of the Hepadnaviridae more closely. The figure shows an outline of HFV replication, with the unique aspects in red text and arrows, and non-unique stages in black. Reverse transcription occurs at a late step in the replication cycle, resulting in infectious HFV particles with DNA not RNA which has raised the question of whether HFV is a DNA or an RNA virus. Incomplete cleavage of the Gag protein by viral protease during maturation means that HFV doesn’t contain matrix, capsid, and nucleocapsid proteins, only two large Gag proteins. Particles bud from cells primarily through the endoplasmic reticulum. HFV’s cellular receptor hasn’t yet been identified, but must be ubiquitous, since HFV has a wide tropism.
Source: Historical Perspective of Foamy Virus Epidemiology and Infection (Meiering et al. Clinical Microbiology Reviews, January 2001)