Reoviruses have three highly interesting and characteristic features
regarding replication. They are dsRNA viruses which replicate in the
cytoplasm (indicating they have everything they need for replication and
do not utilize the cells replication enzymes) and they do not fully uncoat
during the process of replication. The reason for their failure to fully
uncoat is
that the coat is resistant to protease digestion, which prevents them from
being completely destroyed by the infected cell. The mRNA used in
translation is
synthesized from the negative strand of the dsRNA. Each negative strand
produces many positive strands. The process utilizes particle-associated
transcriptase. Neither of the strands of dsRNA appear among the
transcription products, and both strands remain in the uncoated core
particle. The capped positive strand of RNA found in progeny dsRNA is
synthesized from the negative strand template found in the dsRNA within
the parental cores. These capped transcripts that result from primary
transcription are not polyadenylated. Later in infection, secondary
transcription occurs and results in uncapped non-polyadenylated
transcripts. mRNAs leave the core and get translated in the cytoplasm.
The segmented genome allows for possible reassortment, something that has
yet to be seen naturally but can be made to occur in vitro.
Several differences exist in the modes of entry into and exit from cells
between the different genera in the Reoviridae family. The most
extensively studied virus of the Reoviridae family are the Orthreoviruses
which enter the cell either by receptor mediated endocytosis or as a
result of digestion with chymotrypsin in the intestine. Intermediate
subviral particles may pass into the cytoplasm by the endosmal pathway or
directly.
Initially in replication only certain genes are transcribed, the others
are
transcribed after the synthesis of early viral protein. The virions are
released by cell lysis after they have self-assembled and have accumulated
in the inclusion bodies in the cell's cytoplasm.
Since Rotaviruses are the single most important etiologic agents of severe
diarrheal infection in young children, the concentration here will lay
with their cycle of replication. The replication cycle has been primarily
studied in continuous cell cultures made from monkey kidneys. The
cycle in these cells has maximum yields at about 10-12 hours. Key
features of the cycle are:
1)Rotavirus' ligands on the outer capsid hemagglutinating protein VP4 bind
to sialic acid receptors on the cell about to be infected.
2)In order for the virion to enter the cytoplasm, cleavage of VP4 by
trypsin is required. This occurs when the virion makes direct contact
with the cytoplasm.
3)The outer capsid glycoprotein VP7 is retained in the rough endoplasmic
reticulum.
4)The Rotavirus precursor, with the help of a nonstructural glycoprotein,
buds off into the cisternae of the rough endoplasmic reticulum. Here, it
acquires a temporary envelope which is later removed so that the entire
outer capsid can be assembled by VP7.
5) Inclusion bodies are formed in infected cells 6-7 hours after
infection.
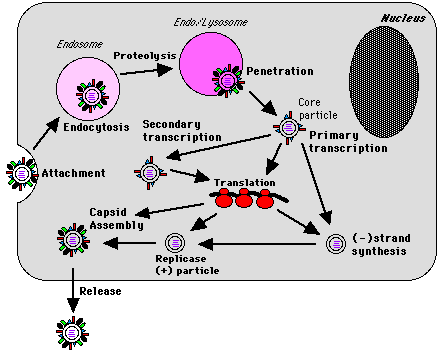 Major features of the rotavirus replication cycle.
Reprinted from
Dr. Alan
Cann's Tulane website
Major features of the rotavirus replication cycle.
Reprinted from
Dr. Alan
Cann's Tulane website
Back

