 |
|
|
 |
 |
 |
 |
 |
 |
 |
Selected media coverage
(some links may require a subscription)- New DNA analysis supports an unrecognized tribe's ancient roots in California (New York Times, April 12, 2022)
- Did we sicken Neanderthals to death? (Haaretz, November 5, 2019)
- Genome hackers show no one's DNA is anonymous any more (Wired, October 11, 2018)
- Genetic privacy and the case of the Golden State Killer — diving into the science (Genetic Literacy Project, May 1, 2018)
- DNA, speech data reveal history of genetic, linguistic evolution of Cape Verdean Creole (O Jornal, March 9, 2018)
- CODIS has more ID information than believed, scientists find (Forensic Magazine, May 15, 2017)
- Thousands of horsemen may have swept into Bronze Age Europe, transforming the local population (Science, February 21, 2017)
- Does CODIS contain untapped ancestry information? (Forensic Magazine, April 13, 2016)
- Human genetic diversity and social inequalities (Genes to Genomes, September 9, 2015)
- How languages and genes evolve together (The Atlantic, February 3, 2015)
- Scholars pursue Jews' genetic trail: Humanities professor, evolutionary biologist team up (J Weekly, May 16, 2013)
- All the variation (Genome Technology, October 2011)
- Researchers uncover unexpected genetic relatedness within HapMap 3 populations (Genome Technology, December 2010/January 2011)
- Genetic mutations offer insights on human diversity (Washington Post, February 22, 2008)
- La gran familia de emigrantes africanos (Público, February 21, 2008)
- Tracing the first Americans (American Museum of Natural History, Hall of Human Origins, December 17, 2007)
- Northwest Passage: Americas populated via Alaska, genetics show (Science News 172: 339, December 1, 2007)
- Early humans settled globe gradually, gene study says (National Geographic News, October 18, 2005)
- Read all about it: The Lancet's Paper of the Year, 2003 (Lancet 362: 2101-2103 [2003])
- Race and gene studies: what difference makes a difference? (California Newsreel, 2003)
- Genes show the way home (The Times [London], December 20, 2002)
- People are same, but different (Los Angeles Times, December 20, 2002)
- Cada persona es distinta, pero no debido
a su raza (El País, December 20, 2002)
Somebody has translated our "Genetic structure of human populations" paper into Spanish: Estructura genética de las poblaciones humanas
Coverage by university news
- Study reveals how cultural factors influence chess move choice (Stanford Report, November 14, 2023)
- Researchers illuminate centuries of identity lost because of slavery (USC News, July 11, 2023)
- A new approach to genetic genealogy sheds light on African American ancestry (Stanford Report, July 10, 2023)
- Stanford researcher applies evolutionary math to March Madness (Stanford Report, March 13, 2023)
- Stanford and Illinois researchers publish genomic evidence of ancient Muwekma Ohlone connection (Stanford Report, March 21, 2022)
- Study ties present-day Native American tribe to ancestors in San Francisco Bay Area (Illinois News Bureau, March 21, 2022)
- Stanford professor spotlights evolutionary tree concepts with campus trees (Stanford Report, December 7, 2021)
- Stanford model reveals surprising disconnect between physical characteristics and genetic ancestry in certain populations (Stanford Report, April 5, 2021)
- Models explore how disease dynamics change when cultural behaviors spread like a pathogen (Stanford Report, September 8, 2020)
- Stanford scientists commemorate 50 years of a "famously obscure" field (Stanford Report, June 1, 2020)
- Formula for an "aha moment": pair up with your spouse (Stanford Medicine Scope, February 13, 2020)
- Deadly human diseases may have killed off the Neanderthals (Berkeley News, November 20, 2019)
- Stanford scientists link Neanderthal extinction to human diseases (Stanford Report, November 7, 2019)
- Stanford researchers discover a new way to find relatives from forensic DNA (Stanford Report, October 17, 2018)
- Cape Verde creole, DNA, speech data reveal history of genetic, linguistic evolution (University of Michigan Global Michigan, August 28, 2017)
- A fish in a pond or a needle in a haystack? DNA tool rasises promise, privacy concerns (University of Michigan Health Lab Report, May 18, 2017)
- Genetic patterns could aid scientists and police, but raise privacy concerns, Stanford scientists say (Stanford Report, May 15, 2017)
- Forensic DNA reveals more than we thought, and that's both good and bad (Stanford Medicine Scope, May 15, 2017)
- The benefit of mathematical models in medicine (Stanford Medicine Scope, February 19, 2015)
- Human dispersal and the evolution of languages show strong link, Stanford biologists find (Stanford Report, January 30, 2015)
- Probing the deep history of human genes and language (Brown University News From Brown, January 19, 2015)
- Noah Rosenberg and Fiorenza Micheli named to endowed chairs (Stanford Department of Biology news, October 10, 2014)
- Stanford humanities and science scholars join forces for groundbreaking research on Jewish genetics (Stanford Report, March 1, 2013)
- Stanford launches new center to advance 'information age of genomics' (Stanford Report, December 3, 2012)
- Where chromosomes agree, Stanford researchers find signatures of human migrations and marriage practices (Stanford Report, August 14, 2012)
- Research bolsters migration theory (Brown Daily Herald, October 25, 2011)
- U-M study reveals surprising lack of genetic diversity in the most widely used human embryonic stem cell lines (University of Michigan News Service, December 16, 2009)
- Native Americans Descended From a Single Ancestral Group, DNA Study Confirms (UC Davis News and Information, April 28, 2009)
- U-M researchers release most detailed global study of genetic variation (University of Michigan News Service, February 20, 2008)
- Gene study adds weight to theory that native people of the Americas arrived in a single main migration across the Bering Strait (University of Michigan Health System Newsroom, November 27, 2007)
- Study of Genetic Traits Makes Progress (USC News, December 21, 2006)
- One small snip from man; one giant leap for the genome (University of Michigan News Service, December 7, 2006)
- All in the Family (USC News, December 19, 2003)
- DNA suggests humans descend from small ancestral population (Stanford Report, May 28, 2003)
- Family Ties (USC News, December 19, 2002)
Comments in scientific journals on research from the lab
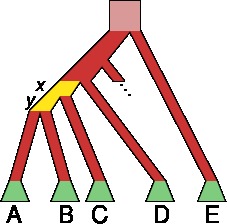
When PhD
student Amy
Goldberg develops a model for sex-biased admixture
on the X-chromosome, a curious mathematical sequence leads
to an unexpected connection deep in the Genetics
archive.
We say goodbye to:
We wish everyone all the best in their new positions!
Zach's thesis on "Human migration, population divergence, and the
accumulation of deleterious alleles: insights from private genetic
variation and whole-exome sequencing" considers several
perspectives on private alleles, including a model of microsatellite
private alleles, a method for counting private alleles in uneven
samples, and a study of connections among rare, private, and deleterious
variants.
Chaolong's thesis on "Statistical methods for analyzing human
genetic variation in diverse populations" considers new
approaches for studying spatial population-genetic variation, and
develops a new method for circumventing allelic dropout in
microsatellite data without requiring replicate genotypes.
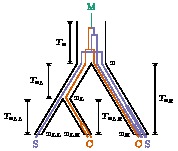
In the
second paper, Laura
Helmkamp and Ethan Jewett have developed three more methods in
the same family of approaches: iSD, iSTEAC,
and iMAC. Laura and Ethan's paper appears in a special issue of
the Journal of Computational Biology in honor of Simon Tavaré
and Mike Waterman.
Past lab news



































 Chaolong, Noah, and Zach at the University of Michigan Department of
Computational Medicine and Biology, with graduate program director
Margit Burmeister and founding center director Gil Omenn
Chaolong, Noah, and Zach at the University of Michigan Department of
Computational Medicine and Biology, with graduate program director
Margit Burmeister and founding center director Gil Omenn


[Washington Post]

A description of distruct appears in Molecular Ecology Notes 4: 137-138 (2004).
