RNA polymerase
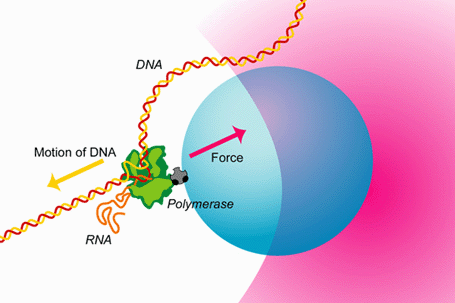
A quick look under a microscope would convince anybody that despite the genetic identity of each of the human body's more than 200 cell types, there is a great morphological diversity between them. This diversity can be accounted for by differences in gene expression patterns in time and space. The central dogma of molecular biology describes the flow of genetic information from DNA (genes) to messenger RNA to enzymatically functional proteins. RNA polymerase (RNAP) is the enzyme responsible for the first step in gene expression—the DNA-directed synthesis of RNA—known as transcription. During transcription RNAP, powered by the free energy released by nucleotide polymerization, can move along the DNA template at speeds greater than 10 nucleotides per second and can support forces greater than 20 pN.
This capacity to sustain large forces explains RNAP's ability to displace bound proteins, such as histones, that it encounters as it transcribes along the DNA template. The ability to convert chemical energy into motion is similar to that of other motor proteins such as myosin and kinesin. However, the fact that RNAP moves along DNA allows it to not only be regulated by extrinsic protein factors, but also by signals encoded within the DNA template. This extra degree of regulation makes RNAP one of the most exquisitely controlled proteins in the cell. The regulation of gene expression can only be understood once we understand the detailed mechanism by which RNAP catalyzes the addition of a nucleotide to a growing RNA strand and couples this to its translocation along the DNA template.
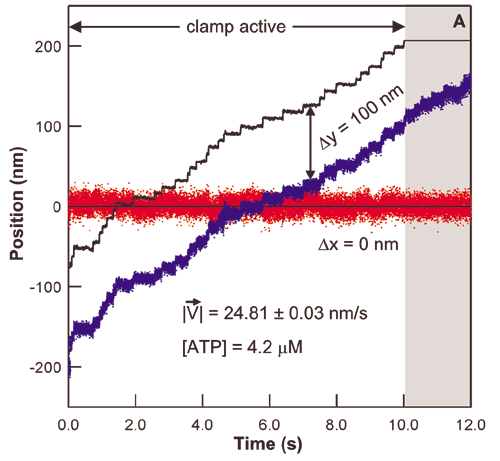
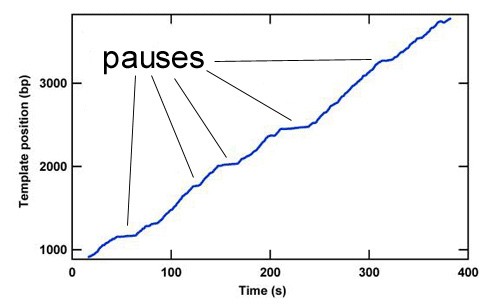
The study of the mechanisms underlying RNAP transcription by conventional biochemical experiments, which report only ensemble averages, are complicated by the rapid loss of synchronization in populations of transcribing RNAP complexes. To overcome this difficulty, the concentration of NTPs is often reduced below physiological levels. Using optical traps we are able to track single transcribing molecules of RNAP at physiological NTP concentrations, thereby directly measuring kinetics and providing accurate spatial and temporal information. The trace below shows RNAP position versus time along the rpoB gene template. Optical trapping allows us to probe low probability short-lived events, such as the pauses in this trace, which would never be seen by traditional biochemical methods.
DNA and RNA structure
DNA and RNA form the fundamental building blocks of the universal genetic code. They can form complex structures which interact with proteins, other nucleic acids, and even small regulatory molecules. Understanding the fundamental properties of these molecules gives us not only important physical insight into the physics of polymers, but also into the role their structures and interactions play in biological systems, from the simplest DNA-DNA interactions all the way up to their role in complex macromolecular machines like RNA polymerase and the ribosome. RNA can even play a role as an enzyme (so-called ribozymes) which can directly catalyze chemical reactions and regulate genetic expression.
In the Block Lab, we are capable of directly measuring and applying forces to nucleic acid polymers using optical trapping. This capability gives us the ability to study them from a unique perspective, one which is complementary to more traditional biochemical approaches. We have used this approach to probe the fundamental thermodynamics of DNA and RNA structures, as well as the biological function of riboswitches. We have also been involved in developing new technologies for combining fluorescence with optical trapping.
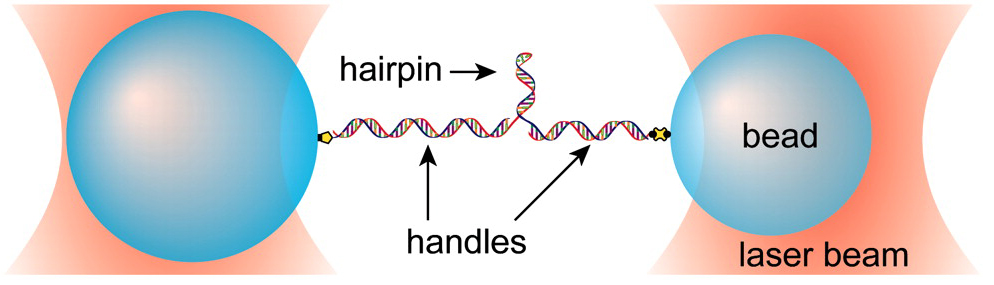
Riboswitches are mRNA elements that regulate gene expression through ligand-induced conformational changes. They typically lie in the 5′ untranslated region of some genes and consist of a ligand binding aptamer domain followed by an expression platform. Aptamer conformation affects the structure adopted by the expression platform (e.g., the formation or absence of a terminator hairpin) and thus the gene's expression. Among the simplest riboswitches are those involved in purine metabolism, which contain aptamers with a three helix junction. We have studied the aptamer of one such purine riboswitch, the pbuE adenine riboswitch. Using our single molecule dumbbell assay we are able to effectively hold the RNA of interest between two beads and exert force on it in order to probe aptamer structure. We measure extension change during folding and unfolding of the aptamer and relate extension changes to numbers of nucleotides and ultimately to specific aptamer structures.
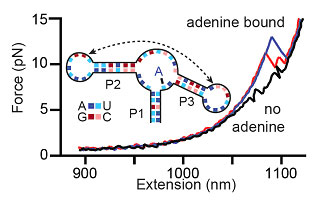
As an example, the figure below shows force-extension curves while unfolding the aptamer. In the presence of adenine (the aptamer's ligand), the RNA is often ligand bound and stabilized; it only rips to the unfolded state at high force. Force-extension curves showing aptamer unfolding. Without adenine, two events are seen (black), corresponding to the unfolding of hairpins P2 and P3 (inset). With adenine bound to the aptamer, large unfolding events are observed (blue), sometimes involving an intermediate state (red), that correspond to opening of the entire aptamer structure. We have extracted energies, distances, and rate constants describing aptamer folding by using force to study aptamer kinetics and equilibrium folding transitions. The adenine-induced stabilization of the closing aptamer helix, P1, was described and a quantitative energy landscape for riboswitch aptamer folding was created, dissecting the secondary and tertiary folding events of this RNA structure.
Kinesin
Kinesin molecules use the energy of ATP hydrolysis to move in discrete steps along protein filaments called microtubules in cells. Each molecule of kinesin consists of two catalytically active motor domains, called "heads" that bind both a microtubule and ATP. The two heads are joined by short polypeptide chain, called the "neck linker" to a long coiled-coil stalk that terminates in a distal cargo binding domain.
A single (dimeric) protein molecule—consisting of only ~800 amino acids, or roughly ten thousand atoms—is sufficient to move long distances along a microtubule, even against considerable load. By studying the motion of single molecules of kinesin, our aim is to understand how this tiny machine works. Historically, most measurements of kinesins have focused on kinesin-1 the founding member of a diverse family of related motor proteins. Kinesin proteins are essential for vesicle transport in neurons, and chromosome movement in dividing cells. By learning how different kinesin proteins work, we take an important step toward comprehending the dynamics of living cells.
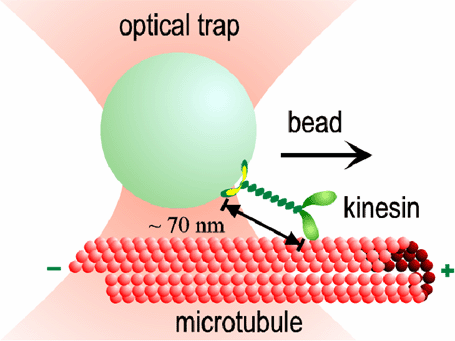
Our optical traps have become very sophisticated, with automatic feedback control that allows us to apply forces of constant magnitude and direction to the kinesin motors while they move. Studying the effect of load on the speed and regularity of kinesin's stepping motion provides clues about how motion is coupled to the biochemical events of ATP hydrolysis. For example, our data suggest that at least 3 nm of each 8 nm step occurs all at once, during a single biochemical transition. This "working stroke" must be well-aligned with the microtubule. The remaining 5 nm may occur simultaneously, or may occur after a slight (<1 ms) delay. We also find evidence for smaller (<1 nm) side-to-side motions occurring at other times in the biochemical cycle.